Animals
Six- to eight-week-old female CD45.2+Thy1.2+ C57BL/6 (C57BL/6J) mice, CD45.1+ mice (B6.SJL-Ptprca Pepcb/BoyCrl) and NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were purchased from Charles River Laboratories. CD45.1+CD45.2+ mice were generated by crossing CD45.1+ mice with CD45.2+ C57BL/6 mice. TCR-transgenic Thy1.1+ pmel-1 (PMEL) mice (B6.Cg-Thy1a/Cy Tg(TcraTcrb)8Rest/J) and TCR-transgenic OT-I mice (C57BL/6-Tg(TcraTcrb)1100Mjb/J), CD45.2+ background Rosa26-Cas9 knock-in mice (B6J.129(Cg)-Gt(ROSA)26Sortm1.1(CAG-cas9*,-EGFP)Fezh/J) were originally purchased from the Jackson Laboratory and maintained at the EPFLâs pathogen-free facility. OT1 mice were crossed with CD45.1+ mice to generate CD45.1+ OT1 mice. CRISPRâCas9 knock-in CD45.2+ mice were crossed with CD45.1+ OT1 mice to generate CRISPRâCas9 knock-in OT1 TCR-transgenic mice. Tcf7DTR-GFP P14 mice on a CD45.2 background were generated as described before9 and maintained at the University of Lausanneâs pathogen-free facility. All mice were housed in the EPFL Center of PhenoGenomics or a conventional animal facility of the University of Lausanne and were kept in individually ventilated cages, at 19â23â°C, with 45â65% humidity and with a 12âh darkâlight cycle. Experimental procedures in mouse studies were approved by the Swiss authorities (Canton of Vaud, animal protocol IDs VD3206, VD3533, VD3902, VD3912, VD3915 and VD3040x2d) and performed in accordance with the guidelines from the Center of PhenoGenomics of the EPFL and the animal facility of the University of Lausanne.
Cell lines and tumour models
B16F10 melanoma cells, MC38 mouse colon adenocarcinoma cells, Raji human lymphoma cells, CTLL-2, K562, human embryonic kidney 293T (HEK293T) and Phoenix-Eco cells were originally procured from the American Type Culture Collection. B16-OVA mouse melanoma cell lines were provided by D.J. Irvine (Massachusetts Institute of Technology). B16-gp33 and YUMM1.7-OVA mouse melanoma cell lines were provided by W. Held (University of Lausanne). HER2-transduced MC38 mouse colon cancer cell lines (MC38-HER2) were provided by P. Romero (University of Lausanne). Luciferase-positive Nalm6 cells (Nalm6-luciferase) were provided by S. Chen (Yale University). All cell lines were confirmed mycoplasma-free before use. All mouse tumour cells, HEK293 cells and Phoenix-Eco cells were cultured in complete DMEM (Thermo Fisher Scientific) supplemented with fetal bovine serum (FBS) (10% v/v, Thermo Fisher Scientific), HEPES (1% v/v, Thermo Fisher Scientific), penicillin/streptomycin (1% v/v, Thermo Fisher Scientific), sodium pyruvate (1% v/v, Thermo Fisher Scientific) and 2-mercaptoethanol (0.1% v/v, Thermo Fisher Scientific). Raji lymphoma cells and Nalm6-luciferase cells were cultured in complete RPMI medium (containing RPMI-1640, FBS (10% v/v), HEPES (pHâ7.2â7.5, 1% v/v), penicillin/streptomycin (1% v/v) and sodium pyruvate (1% v/v)). CTLL-2 cells were cultured in complete RPMI medium (containing RPMI-1640, FBS (10% v/v), HEPES (pHâ7.2â7.5, 1% v/v), penicillin/streptomycin (1% v/v), sodium pyruvate (1% v/v) and 2-mercaptoethanol (0.1% v/v)) supplemented with mouse IL-2 (50ângâmlâ1, PeproTech). B16F10, B16-gp33, B16-OVA, YUMM1.7-OVA, MC38 or MC38-HER2 tumour cells (5âÃâ105, 1âÃâ106 or as indicated) were implanted s.c. into the right flanks of CD45.2+Thy1.2+ C57BL/6 WT mice or CD45.1+CD45.2+ C57BL/6J mice to establish the syngeneic tumour models. Raji lymphoma cells (2âÃâ106) suspended in Matrigel (Corning) were implanted s.c. into the right flanks of NSG mice to establish the Raji lymphoma model. Nalm6-luciferase cells (1âÃâ106) suspended in PBS were i.v. injected into NSG mice to establish the metastatic leukaemia model and the survivor mice from treatment were rechallenged with Nalm6-luciferase cells (1âÃâ106, i.v.) to establish the recurrent leukaemia model. In rechallenge studies, the corresponding tumour cells (1âÃâ105, 5âÃâ105 or as indicated) were s.c. implanted into the left flanks of cured mice 2 or 3âmonths after the initial tumour inoculation.
Production of mouse and human FcâIL-4 proteins
As reported previously, both mouse and human FcâIL-4 fusion proteins were engineered by fusing IL-4 to the C terminus of mutant non-lytic IgG2a Fc35 by a GS4 linker and then expressed by FreeStyle 293-F cells (Thermo Fisher Scientific) at the EPFL Protein Production and Structure Core Facility12. The supernatant of the cell culture medium containing the recombinant protein was filtered through a 0.22âμm membrane (Millipore) to remove cell debris. The recombinant protein was first captured with a HiTrap Protein A affinity chromatography column (Cytiva, 17-0403-01, 5âml) on an AKTA Pure 25 (GE Healthcare) and then eluted with an elution buffer (0.05âM sodium citrate, 0.3âM sodium chloride, pHâ3.3). The eluted protein was collected immediately in a neutralization buffer (1âM Tris HCl, pHâ10.0) and then concentrated using the ultrafiltration method (molecular weight cut-off 10âkDa). The concentrated protein solution was further purified with a Superdex 200 Increase size-exclusion chromatography column (GE Healthcare). The purified protein was aliquoted and stored at â80â°C before use. The purity of the recombinant protein was confirmed with sodium dodecyl sulfateâpolyacrylamide gel electrophoresis (SDSâPAGE) and the bioactivity was compared with that of commercial IL-4 (Biolegend).
Preparation of PMEL, OT1 and Tcf7
DTR-GFP P14 CD8+ T cells
Spleens from PMEL, OT1 or Tcf7DTR-GFP P14 mice were cut into small pieces and then mechanically meshed through a 70âμm strainer (Fisher Scientific) to obtain a single-splenocyte suspension. ACK lysis buffer (2âml per spleen, Thermo Fisher Scientific) was added to the above splenocyte pellets to lyse the red blood cells for 3âmin at room temperature. After washing twice with cold PBS (Thermo Fisher Scientific) and filtering through a 70âμm strainer again, splenocyte pellets were then resuspended at a cell density of 1âÃâ106 per ml in complete RPMI medium (containing RPMI-1640, FBS (10% v/v), HEPES (pHâ7.2â7.5, 1% v/v), penicillin/streptomycin (1% v/v), sodium pyruvate (1% v/v) and 2-mercaptoethanol (0.1% v/v)) supplemented with mouse IL-2 (10ângâmlâ1, PeproTech) and IL-7 (1ângâmlâ1, PeproTech), as well as human gp10025â33, OVA257â264 or LCMV gp33â41 peptide (0.5 or 1âμM, GenScript) for PMEL, OT1 and P14 T cells, respectively. After 2 or 3âdays of culture, live cells were collected by density gradient centrifugation against Ficoll-Paque PLUS (GE Healthcare). The enriched cells were cultured for another 3âdays at a cell density of 0.5âÃâ106 per millilitre in complete RPMI medium supplemented with mouse IL-2 (10ângâmlâ1) and IL-7 (10ângâmlâ1) to obtain activated CD8+ T cells with purity greater than 95% (flow cytometry analyses).
Preparation of HER2-CAR-T cells
The spinoculation method was used for the preparation of HER2-CAR-T cells. Briefly, Phoenix-Eco cells were first transfected with HER2-CAR carrying plasmid and pCL-Eco packaging plasmid using the calcium phosphate method. After refreshing the supernatant 12âh after the transfection, the virus-containing supernatant for T cell transduction was collected every 24âh after the transfection until 72âh. Before the transduction, splenocytes from WT mice were stimulated with coated anti-mouse CD3 antibody (5âµgâmlâ1, 17A2, BioXcell) and soluble anti-mouse CD28 antibody (5âµgâmlâ1, PV-1, BioXcell) in the presence of IL-2 (10ângâmlâ1) for 1âday. The activated CD8+ T cells were then isolated as described above for subsequent spin transduction. The above-collected virus-containing supernatant was dispensed into a non-tissue-culture-treated six-well plate, which was precoated with protamine (10âµgâmlâ1, Sigma-Aldrich) overnight at 4â°C and then blocked with PBS containing FBS (v/v 1%) for 20âmin before use. The plate loaded with virus-containing supernatant was centrifuged at a speed of 2,000g for 2âh at 32â°C to absorb virus particles on the bottom of the plate and the supernatant was then aspirated. Activated CD8+ T cells suspended in the cell culture medium supplemented with mouse IL-2 (10ângâmlâ1) and IL-7 (10ângâmlâ1) were immediately added to the plate and centrifuged at 300g for 15âmin at 32â°C. The transduction was repeated once 24âh later and the cells obtained were cultured for another 2 or 3âdays before use. The transduction efficiency was determined 48âh post-transduction. Untransduced T cells activated by coated anti-mouse CD3 and CD28 antibodies were used as a control.
Preparation of human CD19-CAR-T cells
Primary T lymphocytes from healthy donors were provided by the Cleveland Clinicâs BioRepository Core in accordance with the guidelines from Cleveland Clinicâs BioRepository Review Committee. T cells from healthy donor peripheral blood mononuclear cells were purified by negative selection using a T cell isolation kit (Miltenyi) and subsequently activated with CD3/CD28 Dynabeads (Thermo Fisher Scientific) at a cell-to-bead ratio of 1:3. A lentiviral vector carrying a previously described CD19-specific CAR with 4-1BB/CD3ζ transgene was constructed36 and was used to transduce the cells during the activation phase. It was washed out 3âdays after the initiation of culture37. Cells were cultured in OpTmizer T Cell Expansion Basal Medium (Thermo Fisher Scientific) supplemented with GlutaMAX supplement (2âmM, Thermo Fisher Scientific), human serum AB (5%, Gemini Bioproducts), IL-7 (5ângâmlâ1, Miltenyi) and IL-15 (5ângâmlâ1, Miltenyi). Cell expansion was facilitated using a rocking platform (WAVE Bioreactor System) for a duration of 8 to 12âdays, and the beads were then magnetically removed. Finally, CAR-T cells were collected and cryopreserved until the assays were performed.
Preparation of IL-4Rα-KO OT1, STAT6-KO OT1 and LDHA-KO OT1 CD8+ T cells
To prepare IL-4Rα-KO OT1 (OT1IL-4Rα-KO), STAT6-KO OT1 (OT1STAT6-KO) and LDHA-KO OT1 (OT1LDHA-KO) CD8+ T cells, CRISPRâCas9 knock-in OT1 CD8+ T cells were isolated from splenocytes of Cas9 knock-in OT1 TCR-transgenic mice using a CD8 negative selection kit (Miltenyi Biotec) and activated with coated anti-mouse CD3 antibody (5âµgâmlâ1, 17A2, BioXcell) and soluble anti-mouse CD28 (5âµgâmlâ1, PV-1, BioXcell) antibody in the presence of IL-2 (10ângâmlâ1) for 1âday. The activated CD8+ T cells were then spin-transduced twice on days 2 and 3 with retroviruses containing scrambled control guide RNA (gRNA), IL-4Rα-targeting gRNA, STAT6-targeting gRNA or LDHA-targeting gRNA in a non-tissue-culture-treated six-well plate coated with protamine (10âµgâmlâ1, Sigma-Aldrich) as described above. Transduced CD8+ T cells were then expanded for another 3âdays before in vitro or in vivo use. The pool of gRNAs targeting IL-4Rα (IL-4Rα-1, 5â²âGCAGCAGCGGGGACTGACGAâ3â²; IL-4Rα-2, 5â²âGACACCCTCAAACTTGTCAGâ3â²; and IL-4Rα-3, 5â²â GGCCCCAGTACAGAATGTGGâ3â²), gRNAs targeting STAT6 (STAT6-1, 5â²âCACCGTTGACTTTCCACAACGCCTAâ3â²; STAT6-2 5â²âCACCGAGTTTACTACAGCCCTCGGAâ3â²; and STAT6-3, 5â²âCACCGATAAAGCGCTGTGAGCGGAAâ3â²) gRNAs targeting LDHA (LDHA-1, 5â²âGTTGCAATCTGGATTCAGCGâ3â²; LDHA-2, 5â²âGTCATGGAAGACAAACTCAAâ3â²; and LDHA-3, 5â²âGAAGTCTCTTAACCCAGAACâ3â²) and a scrambled gRNA control (5â²âGCGAGGTATTCGGCTCCGCGâ3â²) were designed using the publicly available online gRNA design tool CRISPick. The knockdown efficiency was evaluated at protein expression levels with flow cytometry or western blot.
Preparation of LDHA-knockdown PMEL CD8+ T cells
To prepare LDHA-knockdown PMEL (PMELLDHA-KD) CD8+ T cells, shLDHA lentivirus particles were produced in HEK293T cells that were transfected with plasmids of pVSV-G, Delta 8.9 and pLKO.1 puro_shLDHA using the calcium phosphate method as described above. PMEL CD8+ T cells were primed for 1âday as described above. Live activated PMEL CD8+ T cells were collected by density gradient centrifugation against Ficoll-Paque PLUS, resuspended in the complete cell culture medium supplemented with mouse IL-2 (10ângâmlâ1) and IL-7 (10ângâmlâ1), and spin-transduced with shLDHA twice, on days 2 and 3. The pool of shLDHA was designed as follows: shLDHA1, 5â²âGTTCCCAGTTAAGTCGTATAATCTCTTGAATTATACGACTTAACTGGGAACTTTTTTGGTACCâ3â²; shLDHA2, 5â²âCGTGAACATCTTCAAGTTCATTCTCTTGAAATGAACTTGAAGATGTTCACGTTTTTTGGTACCâ3â²; shLDHA3, 5â²âCGTCTCCCTGAAGTCTCTTAATCTCTTGAATTAAGAGACTTCAGGGAGACGTTTTTTGGTACCâ3â². The PMELLDHA-KD CD8+ T cells obtained were expanded for another 3âdays before in vitro or in vivo use. The knockdown efficiency was evaluated at gene and protein expression levels.
Preparation of LDHA-overexpression PMEL CD8+ T cells
To prepare LDHA-overexpression PMEL (PMELLDHA-OE) CD8+ T cells, the retroviral particles were produced in Pheonix Eco cells that were transfected with the pMSGV plasmid containing a Thy1.1 reporter and the LDHA gene using the polyethylenimine (PEI) transfection method. Briefly, PEI (80âµg) was mixed with pMSGV plasmid (21.4âµg) and pCL-Eco plasmid (14.4âµg) in a total of 2âml serum-free medium. The mixture was incubated at room temperature for 20âmin, during which the medium in the T150 flask containing the Pheonix Eco cells was aspirated. The solution of the PEIâDNA complex was then added to the Pheonix Eco cells followed by an incubation at room temperature for 5âmin and addition of RPMI medium (14âml). Virus was collected 48âh later and stored at â80â°C until transduction. PMEL CD8+ T cells were isolated and primed in T75 flasks coated with anti-CD3 and anti-CD28 antibodies for 24âh in complete RPMI cell culture medium supplemented with mouse IL-2 (10ângâmlâ1) and IL-7 (10ângâmlâ1), followed by transduction with the LDHA-overexpression retrovirus using the spinoculation method. The LDHA sequence used was National Center for Biotechnology (NCBI) reference sequence NM_010699.2. The PMELLDHA-OE CD8+ T cells obtained were expanded in vitro for another 3âdays. The transduction efficiency was evaluated at gene and protein expression levels before use.
Collection of tumour-infiltrating immune cells for analyses
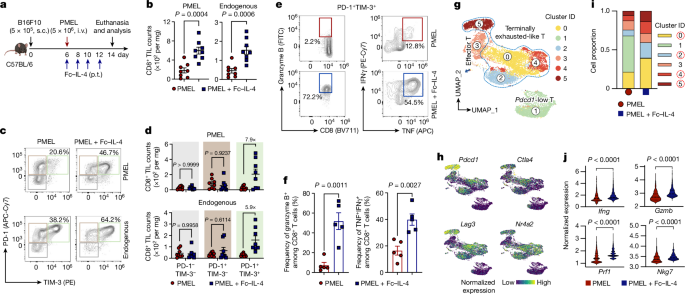
CD45.2+Thy1.2+ C57BL/6 mice bearing B16F10 tumours received i.v. adoptive transfer of activated PMEL CD8+ T cells (5âÃâ106 per mouse), followed by p.t. administration of FcâIL-4 (20âμg per mouse) or PBS control every other day for four doses. For the BrdU experiments, mice were intraperitoneally (i.p.) administered BrdU (1âmg per mouse, Sigma-Aldrich) 1âday before tumour tissue collection. Tumours were collected, weighed, mechanically minced and stirred at 1,000âr.p.m. in RPMI-1640 medium with collagenase type IV (1âmgâmlâ1, Thermo Fisher Scientific), dispase 2 (100âμgâmlâ1, Sigma-Aldrich), hyaluronidase (100âμgâmlâ1, Sigma-Aldrich) and DNase I (100âμgâmlâ1, Sigma-Aldrich) for 60âmin at 37â°C for digestion. Red blood cells in the digested tumour samples were lysed with ACK lysing buffer for 3âmin at room temperature. Tumour-infiltrating leukocytes were then enriched by density gradient centrifugation against Percoll (GE Healthcare), resuspended in PBS with bovine serum albumin (0.2%, w/v, Sigma-Aldrich), stained with the indicated antibodies and analysed by flow cytometry.
Flow cytometry analyses
For surface marker staining, cells were first blocked with anti-mouse CD16/32 antibodies (BioLegend) and incubated with the indicated antibodies at 4â°C for 15âmin, followed by live/dead staining using 4,6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich) or Zombie Aqua Fixable Dye (BioLegend). Cells were then washed with PBS containing bovine serum albumin (0.2%, w/v) and resuspended in the same buffer for flow cytometry analyses. For intracellular cytokine staining, cells were first stimulated with Cell Stimulation Cocktail (protein transport inhibitors included, Invitrogen/Thermo Fisher Scientific) at 37â°C for 4â6âh. Cells were then processed as for surface marker staining, followed by live/dead staining using Zombie Aqua Fixable Dye. Next, cells were fixed and permeabilized with the Cytofix/Cytoperm Fixation/Permeabilization Solution Kit (BD Biosciences) according to the manufacturerâs instructions, followed by incubation with indicated antibodies for intracellular cytokine staining. Intracellular active caspase-3 staining was conducted similarly, except that pre-stimulation with the Cell Stimulation Cocktail was not applied. For BrdU and transcription factor staining, cells were first stained for surface markers and with Zombie Aqua Fixable Dye as described above. Next, cells were fixed and permeabilized with the Foxp3/Transcription Factor Staining Buffer Set (eBioscience) according to the manufacturerâs instructions, followed by incubation with the indicated antibodies. For intracellular phosphorylated protein staining, cells were first stained for surface markers and with Zombie Aqua Fixable Dye as described above. Next, cells were fixed with a solution of paraformaldehyde (1.5%) at room temperature for 15âmin and permeabilized with chilled methanol at 0â°C for 10âmin. On complete removal of the paraformaldehyde solution and methanol, cells were incubated with the indicated antibodies for intracellular phosphorylated protein staining at room temperature for 30âmin. Data were collected using an Attune NxT Flow Cytometer with Attune NxT Software v.3 (Invitrogen/Thermal Fischer Scientific). Analyses were performed using FlowJo v.10.6.1 (Tree Star). Gate margins were determined by isotype controls and fluorescence-minus-one controls.
Antibodies and reagents for flow cytometry and western blot
The following antibodies or staining reagents were purchased from BioLegend: CD16/32 (93, 101302), Thy1.1 (OX-7, 202529), Thy1.2 (30-H12, 105343), CD45.1 (A20, 110707), CD45.2 (104, 109814), CD8α (53-6.7, 100714), CD8β (YTS256.7.7, 126606), CD4 (RM4-5, 100526), NK1.1 (PK136, 108740), F4/80 (BM8,123108), CD3ε (17A2, 100306), CD19 (6D5,115520), CD44 (IM7, 103006), CD11c (N418, 117348), I-A/I-E (MHC-II, M5/114.15.2, 107643), Siglec-F (S17007L, 155508), CD80 (16-10A1, 104734), CD86 (GL-1, 105006), Foxp3 (MF-14, 126406), CD11b (M1/70, 101228), granzyme B (GB11, 515403), IFNγ (XMG1.2, 505826), TNF-α (MP6-XT22, 506308), IL-2 (JES6-5H4, 503822), IL-4Rα (I015F8, 144806), CD69 (H1.2F3, 104512), Gr-1 (RB6-8C5, 108423), CD107a (1D4B, 121626), CD95 (SA367H8, 152608), CD178 (MFL3, 106605), PD-1 (29F.1A12, 135216), TIM-3 (RMT3-23, 119706), TIM-3 (RMT3-23, 119737), HRP-actin (2F1-1, 643808), GATA3 (16E10A23, 653805), Tbet (4B10, 644827), anti-rabbit IgG (minimal cross-reactivity) antibody (Poly4064, 406414), anti-mouse IgG1 antibody (RMG1-1, 406617), STAT6 (16G12A08, 657902), Zombie Aqua Fixable Viability Kit (423102), human CD3 (OKT3, 317306), human CD4 (OKT4, 317416), human CD8 (SK1, 344724), human IFNγ (B27, 506516) and human TNF-α (MAb11, 502940). Anti-TCF-7/TCF-1(S33-966, 566693), anti-phospho-Akt (pT308) (J1-223.371, 558275), anti-phospho-Akt (pS473) (M89-61, 560404), anti-Akt (7/Akt/PKBα, 610836) and anti-active caspase-3 (C92-605.rMAb, 570334) were purchased from BD Biosciences. Anti-CD8 (YTS 169.4, BE0117), anti-CD4 (YTS 177, BE0288), anti-NK1.1 (PK136, BP0036), anti-Ly6G (NIMP-R14, BE0320), IgG (LTF-2, BP0090), anti-IL-4 (11B11, BE0045), anti-mouse CD3 (17A2, BE0002), anti-mouse CD28 (PV-1, BE0015-5), anti-mouse PD-1 (RMP1-14, BE0146) and anti-mouse CTLA-4 (9H10, BE0131) were purchased from BioXcell. Goat anti-rat IgG Fc secondary antibody (31226), eBioscience Cell Stimulation Cocktail (00-4970-03) and anti-phospho-STAT6 (Tyr641) (46H1L12, 700247) were purchased from Invitrogen. Anti-rabbit HRP-IgG (7074) and anti-Glut-1 (73015) were purchased from Cell Signalling Technology. Anti-P70S6K (14485-1-AP) and anti-LDHA (19987-1-AP) were purchased from Proteintech. Anti-phospho-P70S6K (pThr389) (ABIN7265266) was purchased from Antibodies-online. Anti-FMC63 scFv (CAR19) (FM3-HPY53) was purchased from ACRO Biosystems. Antibodies for surface staining were used at a 1:100 dilution, for intracellular staining at a 1:50 dilution and for western blot at a 1:1,000 dilution.
scRNA-seq library preparation and sequencing
CD45.2+Thy1.2+ C57BL/6 mice bearing B16F10 tumours received i.v. adoptive transfer of activated Thy1.1+ PMEL CD8+ T cells (5âÃâ106 per mouse), followed by p.t. administration of FcâIL-4 (20âμg per mouse) or PBS control every other day for four doses. Tumours were collected and digested, and tumour-infiltrating PMEL T cells were enriched and sorted with flow cytometry. Sorted T cells were subjected to single-cell isolation and scRNA-seq library building using GEXSCOPE Single Cell RNA Library Kit Cell V2 reagents (Singleron Biotechnologies GmbH) according to the manufacturerâs instructions. Briefly, the cell suspension was loaded into the microfluidic chip and cells were allowed to settle into the microwells. Following the removal of cells that did not settle into the wells, beads containing cell identifying tags (cell barcodes) and unique molecular identifiers (UMI) were flowed into the chip and allowed to settle in the wells on top of the cells. The cells were then lysed and the messenger RNA (mRNA) from each cell hybridized to the barcode sequences on the bead in the same well. After retrieving the beads, the hybridized mRNA was reverse-transcribed into complementary DNA (cDNA). The resulting cDNA was amplified by a minimal number of PCR steps, fragmented, ligated to adaptors and PCR amplified to construct a sequencing library. The gene expression libraries generated were sequenced using an Illumina HiSeq 4000 with a sequencing depth of 50,000 paired-end reads per cell.
scRNA-seq data processing
The raw fastq files were generated and de-multiplexed by CeleScope rna from Singleron (v.3.0.1) and primary data analysis was performed with CeleScope (v.1.10.0) using a custom reference package based on the reference genome (Mus_musculus_ensembl_92). Downstream data analysis was performed with the Seurat v.4 pipeline38. Cells were first filtered on the basis of two metrics: (1) the number of genes detected per cell must be between 200 and 5,000 and (2) the proportion of mitochondrial gene counts (UMIs from mitochondrial genes/total UMIs) must be less than 10%. Next, the gene expression data were normalized using Seurat sctransform39. No major batch effects were observed between the two samples. Finally, the SCT data assay was reduced to two dimensions using uniform manifold approximation and projection (UMAP) for visualization, with 30 computed PCs as input. DEGs were identified using the function âFindMarkersâ for pairwise comparison between two conditions. A log fold-change threshold of 0.25 was applied to select genes as differentially expressed. The function âAddModuleScoreâ was used to calculate the module scores of each cluster on the basis of the aggregated expression of defined gene sets. To identify metabolic subpopulations using scRNA-seq data, we performed an unsupervised clustering analysis and UMAP visualization based on the 1,667 genes involved in KEGG-defined metabolic pathways listed in Supplementary Table 2.
Signalling pathway and upstream regulator analysis
Ingenuity pathway analysis (Qiagen)40 was used to reveal the underlying signalling pathways regulated by the DEGs distinguishing each identified cluster or condition. The DEG list and the corresponding fold-change value, P value and adjusted P value of each gene were loaded into the dataset. The Ingenuity Knowledge Base (genes only) was used as a reference set to perform core expression analysis. T cell-related signalling was selected from the identified canonical pathways to represent the major functional profile of each group. The z score was used to determine activation or inhibition level of specific pathways. Conceptually, the z score is a statistical measure of how closely the actual expression pattern of molecules in our DEG dataset compares to the pattern that is expected on the basis of the literature for a particular annotation. zâ>â0, activated/upregulated; zâ<â0, inhibited/downregulated; zââ¥â2 or zââ¤ââ2 can be considered significant. The P value of each identified signalling pathway was calculated by a right-tailed Fisherâs exact test. The significance indicates the probability of association of molecules from the scRNA-seq dataset with the canonical pathway reference dataset. Ingenuity pathway upstream regulator analysis was used to identify upstream regulators, which refers to any molecules that can affect the expression, transcription or phosphorylation of another molecule, and predict whether they are activated or inhibited given the observed gene expression changes in our DEG dataset. The analysis examines the known targets of each upstream regulator in our dataset, compares the targetsâ actual direction of change to expectations derived from the literature compiled in the Ingenuity Knowledge Base, and then issues a prediction for each upstream regulator.
Ex vivo-induced CD8+ TTE cells
Activated PMEL CD8+ T cells in resting phase (day 6 or 7 in culture after collection from spleens) were re-stimulated with dimeric anti-CD3 antibody (0.5âμgâmlâ1 of anti-CD3 antibody or as indicated), which was prepared by mixing anti-CD3 antibody (17A2, BioXcell) and goat anti-rat IgG (Invitrogen/Thermo Fisher Scientific) at a molar ratio of 2:1, in complete RPMI medium supplemented with IL-2 (10ângâmlâ1) and IL-7 (10ângâmlâ1) for 2âdays. Collected cells were phenotyped with flow cytometry on the basis of the expression level of surface inhibitory receptors (PD-1 and TIM-3). The PD-1+TIM-3+CD8+ T cell subset was sorted as ex vivo-induced CD8+ TTE cells for in vitro and in vivo use.
In vitro coculture of PMEL T cells and tumour cells
B16F10 tumour cells were cultured in complete DMEM as described above. B16F10 tumour cells (1.6âÃâ105 per well) were seeded in a six-well plate at 37â°C overnight. Following aspiration of tumour culture medium, activated PMEL CD8+ T cells were added to the tumour cell culture at an effector/target (E/T) ratio of 1:2 in the presence or absence of FcâIL-4 (20ângâmlâ1). All cells were collected for flow cytometry analyses after 48âh or the indicated time. To determine the lysis of target cells, the cell viability of tumour cells from the coculture was measured with DAPI staining and flow cytometry. Tumour cells cultured alone or with cytokine only, or PMEL CD8+ T cells only, served as controls.
In vitro coculture of human CD19-CAR-T cells and tumour cells
Human CAR-T cells generated as described above were thawed and suspended in FBS-free medium for 2âh followed by positive purification for CD19-CAR-T cells using a magnetic cell separation kit (Miltenyi Biotec) according to the manufacturerâs instructions. Purified CD19-CAR positive CAR-T cells were re-stimulated with K562 cells (E/Tâ=â1/10) for 2âweeks before in vitro use. In a coculture assay, CD19-CAR-T cells were cocultured with Nalm6-luciferase cells (E/Tâ=â1/8) in the presence or absence of human FcâIL-4 (20ângâmlâ1). All cells were collected for flow cytometry analyses after 4âdays. To determine the lysis of target cells, the cell viability of tumour cells from the coculture was measured with DAPI staining and flow cytometry. Tumour cells cultured alone or CAR-T cells only served as controls.
Seahorse assay
Seahorse assay was performed to measure the OCR and ECAR of T cells. Ex vivo-induced CD8+ TTE cells (3âÃâ105 per well) with different treatment conditions were seeded in a Seahorse culture plate (Seahorse Bioscience) in a non-CO2 incubator at 37â°C for 40âmin. OCR and ECAR were measured by an XF96 Seahorse Extracellular Flux Analyzer (Seahorse Bioscience) following the manufacturerâs instructions. In a typical Seahorse assay, cells were treated with oligomycin (1âμM, Sigma-Aldrich), carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP, 1âμM, Sigma-Aldrich), rotenone (0.5âμM, Sigma-Aldrich), antimycin A (0.5âμM, Sigma-Aldrich), glucose (10âmM, Sigma-Aldrich) and 2-DG (50âmM, Sigma-Aldrich). Each condition was performed with 3â6 replicates in a single experiment.
Assay of single-cell ATACâ+âgene coprofiling of ex vivo-induced CD8+ TTE cells
Single-cell coprofiling of epigenomic landscape and gene expression in the same single nuclei was performed using the Chromium Next GEM Single Cell Multiome ATACâ+âGene Expression kit (10x Genomics). Initially, ex vivo-induced CD8+ TTE cells in the presence or absence of IL-4 (20ângâmlâ1, Biolegend) underwent washing, counting and nuclei isolation, with an optimized lysis time of 3âmin. Subsequently, isolated nuclear suspensions were incubated in a transposition mix containing a transposase enzyme, facilitating preferential fragmentation of DNA in open chromatin regions while introducing adaptor sequences to DNA fragment ends. Roughly 9,250 nuclei were loaded onto a Chromium Next GEM Chip J to target a final recovery of roughly 6,000 nuclei. During GEM generation, gel beads introduced a poly(dT) sequence for barcoded, full-length cDNA production from mRNA for gene expression profiling, along with a spacer sequence for barcode attachment to transposed DNA fragments for ATAC profiling. Following GEM incubation, purification and pre-amplification PCR, separate ATAC and gene libraries were constructed using the standard protocol. Both libraries underwent quality assessment before paired-end reads of 150âbp sequencing on an Illumina NovaSeq 6000 sequencing system.
Data processing and analysis for the single-cell ATACâ+âgene coprofiling
The Cell Ranger ARC v.2.0.2 (10x Genomics) was used to perform sample de-multiplexing, barcode processing, identification of open chromatin regions and simultaneous counting of transcripts and peak accessibility in single cells from the sequenced data. Output matrices per barcode underwent joint RNA and ATAC analysis using Signac v.1.12.0 (ref. 41) and Seurat v.4 (ref. 38). Per-cell quality control metrics, including nucleosome banding pattern (stored as nucleosome_signal) and transcriptional start site enrichment score for the ATAC component, were computed, with cells retained on the basis of default settings. Subsequently, peak identification accuracy was enhanced using MACS2 (ref. 42) with the âCallPeaksâ function. We constructed a joint neighbour graph representing both gene expression and DNA accessibility measurements using weighted nearest neighbour methods in Seurat v.4 and investigated potential regulatory elements for genes of interest using the âLinkPeaksâ function. To prepare for motif analyses, we integrated DNA sequence motif information into the dataset using the âAddMotifsâ function and computed a per-cell motif activity score using chromVAR43.
Treatment with pharmacological inhibitors
Ex vivo-induced CD8+ TTE cells were re-stimulated with dimeric anti-CD3 antibody (0.5âμgâmlâ1 or as indicated) in complete RPMI medium supplemented with IL-2 (10ângâmlâ1) and indicated inhibitors (2-DG, 10âmM; FX11, 16âμM; HY-10355, 1âµM; rapamycin, 100ânM; AS1517499, 50ânM or STAT5i, Bestellnummer 573108, 25âµM) for 48âh or as indicated in the presence or absence of FcâIL-4 (20ângâmlâ1). Counts and markers for effector function of live CD8+ T cells were determined by flow cytometry analyses.
Antitumour therapy and rechallenge experiments
Mice bearing established tumours with a size of roughly 25â60âmm2 (day 6 post-inoculation or as indicated) were treated with adoptive transfer of activated PMEL CD8+ T cells, OT1 CD8+ T cells, HER2-CAR-T cells or CD19-CAR-T cells (5âÃâ106 per mouse or as indicated), followed by p.t. administration of FcâIL-4 (20âμg per mouse) or PBS control every other day or as indicated starting from day 6 (four or eight doses in total as indicated). Mice receiving p.t. administration of PBS control only, FcâIL-4 only or untransduced T cells served as controls. For therapy studies with immune checkpoint inhibitors, mice bearing established MC38 tumours with a size of roughly 25âmm2 (day 6 post-inoculation) were treated with an i.p. injection of anti-PD-1 (RMP1-14, BioXcell, 100âµg per mouse) and anti-CTLA-4 (9H10, BioXcell, 100âµg per mouse) antibodies followed by p.t. administration of FcâIL-4 (20âμg per mouse) or PBS control every other day starting from day 6 for four doses in total. Mice receiving PBS only, FcâIL-4 only or anti-PD-1 plus anti-CTLA-4 served as controls. Tumour area and body weight were measured every other day. Tumour area was calculated by the formula areaâ=âlengthâÃâwidth from calliper measurements of two orthogonal diameters. Mice were euthanized when body weight loss was beyond 15%, the tumour area reached 150âmm2 (as a predetermined endpoint) or other endpoints reached the requirements of the animal licences. In the rechallenge studies, B16F10 (1âÃâ105 per mouse), YUMM1.7-OVA (5âÃâ105 per mouse), MC38 (1âÃâ105 per mouse) or MC38-HER2 (1âÃâ105 per mouse) cells were s.c. implanted into the left flanks of cured mice from treatment groups on day 60 or 90 after primary tumour inoculation. Age-matched naive WT mice were s.c. inoculated with the same number of tumour cells as the control. The survival of rechallenged mice was monitored for at least another 60âdays. For the treatment of the recurrent Nalm6 leukaemia model, mice bearing metastatic Nalm6-luciferase tumours were treated with adoptive transfer of CD19-CAR-T cells (2âÃâ106 per mouse) followed by i.p. administration of human FcâIL-4 (100âng per mouse) or PBS control on day 7 and 11 (two doses in total). The survivor mice were i.v. rechallenged with Nalm6-luciferase (1âÃâ106 per mouse) 17âdays after the CAR-T infusion to mimic recurrent leukaemia. The tumour-bearing mice received i.p. administration of d-luciferin potassium salt (Abcam, 150âmgâkgâ1, per mouse) to monitor tumour burden through the IVIS Spectrum In Vivo Imaging System (PerkinElmer). Mice bearing tumours were randomized into groups before treament.Â
Measurement of liver enzymes
CD45.2+Thy1.2+ C57BL/6 mice bearing B16F10 tumours received i.v. adoptive transfer of activated PMEL CD8+ T cells (5âÃâ106 per mouse), followed by p.t. administration of FcâIL-4 (20âμg per mouse) or PBS control every other day for four doses. On day 14, mice were killed and serum samples were collected for analysis. The levels of ALT and AST in serum were measured using Stanbio Chemistry Reagents (Stanbio) according to the manufacturerâs instructions. Healthy mice of the same age without tumours were used as a negative control.
Immune cell depletion study
Mice bearing established B16F10 tumours with a size of roughly 25âmm2 (day 6 post-inoculation) received adoptive transfer of activated PMEL CD8+ T cells (5âÃâ106 per mouse), followed by p.t. administration of FcâIL-4 (20âμg per mouse) as described above. Mice were i.p. injected with the anti-CD8 (YTS 169.4, BioXcell, 400âµg per mouse), anti-CD4 (YTS 177, BioXcell, 400âµg per mouse), anti-NK1.1 (PK136, BioXcell, 400âµg per mouse), anti-Ly6G (NIMP-R14, BioXcell, 400âµg per mouse) or lgG (LTF-2, BioXcell, 400âµg per mouse) antibodies 1âday before the treatment and received another two injections of the antibodies during the treatment to deplete the corresponding immune cells. Tumour area and body weight were measured every other day.
Blockade of T cell egress
Mice bearing B16F10 tumours were treated with adoptive transfer of CD90.1+ PMEL T cells (5âÃâ106âT cells per mouse) followed by administration of FcâIL-4 (20âµg, p.t.) every other day for six doses in total and FTY720 (40âµg, i.p.) every day for nine doses in total to inhibit T cell egress from peripheral lymphoid organs44. Blood was collected every 3âdays to monitor peripheral lymphocytes, and mice were euthanized on day 16 to collect tumour tissues for analysis by flow cytometry.
Depletion of progenitor exhausted CD8+ T cells in vivo
Progenitor exhausted CD8+ T cells were deleted as previously reported9. Briefly, CD45.1+CD45.2+ C57BL/6 mice bearing established B16-gp33 tumours with a size of roughly 25â40âmm2 (day 6 post-inoculation or as indicated) received adoptive transfer of activated CD45.2+ Tcf7DTR-GFP transgenic P14 CD8+ T cells 1âday after whole-body irradiation (4âGy) for lymphodepletion. Diphtheria toxin (1âµg per mouse, Sigma-Aldrich) was i.p. injected twice to deplete the DTR-expressing cells before starting the FcâIL-4 treatments.
Cotransfer of naive OT1 and PMEL T cells
CD45.2+ C57BL/6 mice were sublethally lymphodepleted (4âGy) on day â4 and received adoptive cotransfer of CD45.1+ naive OT1 T cells (2âÃâ106, i.v.) and CD90.1+ naive PMEL T cells (2âÃâ106, i.v.) on day â3. The mice were then inoculated with B16F10 tumour cells on day 0. On day 7, the mice were treated with ACT of activated CD90.2+ PMEL T cells (5âÃâ106, i.v.), followed by administration of FcâIL-4 (20âµg, p.t.) or PBS every other day for four doses. On day 15, mice were euthanized and tumour tissues were collected for flow cytometry analysis.
Adoptive transfer of sorted PD-1+TIM-3â and PD-1+TIM-3+ CD8+ T cells
The PD-1+TIM-3â and PD-1+TIM-3+CD8+ T cell subsets were generated and sorted from ex vivo-induced CD8+ TTE cells as described above. Mice bearing B16F10 tumours received adoptive transfer of PD-1+TIM-3â (1âÃâ106, i.v.) or PD-1+TIM-3+ PMEL T cells (1âÃâ106, i.v.) 1âday after lymphodepletion, followed by treatment with FcâIL-4 (20âµg, p.t.) or PBS every other day for four doses in total. Mice were euthanized on day 15 and tumour tissues were collected for analysis by flow cytometry.
Neutralization of endogenous IL-4
CD45.2+Thy1.2+ C57BL/6 mice bearing B16F10 tumours received i.v. adoptive transfer of activated PMEL CD8+ T cells (5âÃâ106 per mouse), followed by administration of anti-IL-4 antibody (11B11, BioXcell, 200âμg, p.t.) or PBS control every other day for six doses. Tumour area was measured every other day and tumours were collected for analysis of tumour-infiltrating immune cells as described above.
RNA extraction and real-time PCR analysis
Total RNA was extracted using Trizol reagent (Invitrogen) and reverse-transcribed into cDNA using the High-Capacity RNA-to-cDNA kit (Applied Biosystems). Real-time PCR was performed using SYBR Green Master Mix reagents (Applied Biosystems) in the QuantStudio 6 Flex quantitative PCR system. The expression of each gene was calculated on the basis of the cycle threshold, set within the linear range of DNA amplification. The relative expression was calculated by the cycle threshold method, with normalization of raw data to the housekeeping gene Actb.
Metabolomic analysis
Ex vivo-induced CD8+ TTE cells were re-stimulated with dimeric anti-CD3 antibody (0.5âμgâmlâ1) in complete RPMI medium supplemented with IL-2 (10ângâmlâ1) for 48âh in the presence or absence of FcâIL-4 (20ângâmlâ1), and then live CD8+ T cells were sorted for metabolomic analyses. Samples (0.5âÃâ106 CD8+ T cells in 50 µl PBS) were pre-extracted and homogenized by the addition of 200âµl of methanol in the Cryolys Precellys 24 sample homogenizer (twice forâ20âs at 10,000âr.p.m., Bertin Technologies) with ceramic beads. The bead beater was air-cooled at a flow rate of 110âLâminâ1 at 6âbar. Homogenized extracts were centrifuged for 15âmin at 4,000g at 4â°C (Hermle). The resulting supernatant was collected and evaporated to dryness in a vacuum concentrator (LabConco). Dried sample extracts were resuspended in methanol:H2O (4:1, v/v) according to the total protein content. The protein pellets were evaporated and lysed in 20âmM Tris HCl (pHâ7.5), 4âM guanidine hydrochloride, 150âmM NaCl, 1âmM Na2EDTA, 1âmM EGTA, 1% Triton, 2.5âmM sodium pyrophosphate, 1âmM β-glycerophosphate, 1âmM Na3VO4, 1âµgâmlâ1 leupeptin using the Cryolys Precellys 24 sample homogenizer (2âÃâ20âs at 10,000âr.p.m., Bertin Technologies) with ceramic beads. The BCA Protein Assay Kit (Thermo Scientific) was used to measure (by absorbance at 562ânm) total protein concentration (Hidex). Extracted samples were analysed by hydrophilic interaction liquid chromatography coupled to tandem mass spectrometry in both positive and negative ionization modes using a 6495 Triple Quadrupole system interfaced with a 1290 UHPLC system (Agilent Technologies). Pooled quality control samples were analysed periodically throughout the overall analytical run to assess the quality of the data, correct signal intensity drift and remove peaks with poor reproducibility. In addition, a series of diluted quality controls were prepared by dilution with methanol: 100, 50, 25, 12.5 and 6.25% quality controls. Next, metabolites were selected also considering the linear response on the diluted quality control series. Raw liquid chromatography with tandem mass spectrometry data were processed using the Agilent Quantitative analysis software (v.B.07.00, MassHunter, Agilent Technologies). Relative quantification of metabolites was based on extracted ion chromatography areas for the multiple reaction monitoring transitions. The tables obtained (containing peak areas of detected metabolites in all samples) were exported to R software (http://cran.r-project.org/). Signal intensity drift correction and noise filtering was done within the MRM PROBS software. The preprocessed data with peak areas were imported into Metaboanalyst v.5.0 for further data analysis.
Glucose uptake assay
Ex vivo-induced CD8+ TTE cells were re-stimulated with dimeric anti-CD3 antibody (0.5âμgâmlâ1) in complete RPMI medium supplemented with IL-2 (10ângâmlâ1) for 48âh in the presence or absence of FcâIL-4 (20ângâmlâ1). The supernatant was collected to measure the glucose concentration using the Glucose Colorimetric Detection Kit (Invitrogen, EIAGLUC) according to the manufacturerâs instructions. The glucose uptake capacity was calculated with the equation below:
$$\begin{array}{l}{\rm{Glucose}}\;{\rm{uptake}}\;{\rm{capacity}}\\ \,\,\,=\,(1-{\rm{supernatant}}\;{\rm{glucose}}\;{\rm{concentration}}/\\ \,\,\,\,{\rm{fresh}}\;{\rm{medium}}\;{\rm{glucose}}\;{\rm{concentration}})\times 100 \% \end{array}$$
Measurement of cellular NAD+ and NADH
Ex vivo-induced CD8+ TTE cells were re-stimulated with dimeric anti-CD3 antibody (0.5âμgâmlâ1) in complete RPMI medium supplemented with IL-2 (10ângâmlâ1) for 48âh in the presence or absence of FcâIL-4 (20ângâmlâ1). The cellular NAD+ and NADH levels of CD8+ TTE cells were measured using a NAD+/NADH assay kit (Sigma-Aldrich, MAK460) according to the manufacturerâs instructions.
Nicotinamide riboside supplementation assay
Ex vivo-induced CD8+ TTE cells were re-stimulated with dimeric anti-CD3 antibody (0.5âμgâmlâ1) or cocultured with B16F10 at an E/T ratio of 0.5 in the complete RPMI medium supplemented with IL-2 (10ângâmlâ1) for 48âh in the presence or absence of the NAD+ precursor nicotinamide riboside (100âµM). The cell viability, effector function and metabolic activity of CD8+ T cells were measured using flow cytometry and Seahorse assay as described above.
Statistical analysis
Unless stated otherwise, all statistical analyses were performed using GraphPad Prism v.10 (GraphPad Software). Data are presented as meanâ±âs.e.m. unless otherwise specified. Comparison of two groups was performed using a two-sided unpaired Studentâs t-test or MannâWhitney test unless otherwise noted. Comparison of three or more groups was performed using one-way ANOVA with Tukeyâs test or two-way ANOVA and Sidakâs multiple comparisons test unless otherwise indicated. Survival data were analysed using the log-rank test. No statistically significant differences were considered to be present when P values were larger than 0.05. No statistical methods were used to predetermine sample size.
Ethics statement
Experiments and handling of mice were conducted under federal, state and local guidelines and with approval from the Swiss authorities (Canton of Vaud, animal protocol IDs VD3206, VD3533, VD3902, VD3912, VD3915 and VD3040x2d) and performed in accordance with the guidelines of Center of PhenoGenomics at EPFL and the animal facility of the University of Lausanne. Primary T lymphocytes from healthy donors were provided by the Cleveland Clinicâs BioRepository Core in accordance with guidelines from Cleveland Clinicâs BioRepository Review Committee.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.